Overview
This script shows how to generate plot for evaluation metrics of simPIC.
PBMC10k data
Read PBMC10k per-cell-type metric tables from local files.
Show code
pbmc10k_cd4memory <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_cd4memory.txt", sep = " ",header =T, row.names = 1)
pbmc10k_cd4naive <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_cd4naive.txt", sep = " ",header =T, row.names = 1)
pbmc10k_cd8effector <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_cd8eff.txt", sep = " ",header =T, row.names = 1)
pbmc10k_cd14mono <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_cd14mono.txt", sep = " ",header =T, row.names = 1)
pbmc10k_cd8naive <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_cd8naive.txt", sep = " ",header =T, row.names = 1)
pbmc10k_bcell <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_bcellpro.txt", sep = " ",header =T, row.names = 1)
pbmc10k_nkdim <- read.table("~/Downloads/metric_tables_celltypewise/pbmc10k_nkdim.txt", sep = " ",header =T, row.names = 1)Satpathy data
Read Satpathy data per-cell-type metric tables from local files.
Show code
satpathy_monocytes<- read.table("~/Downloads/metric_tables_celltypewise/satpathy_monocytes.txt", sep = " ",header =T, row.names = 1)
satpathy_naivecd8tcell <- read.table("~/Downloads/metric_tables_celltypewise/satpathy_cd8naive.txt", sep = " ",header =T, row.names = 1)
satpathy_regtcell <- read.table("~/Downloads/metric_tables_celltypewise/satpathy_regtcell.txt", sep = " ",header =T, row.names = 1)
satpathy_cd34 <- read.table("~/Downloads/metric_tables_celltypewise/satpathy_cd34progenitor.txt", sep = " ", header =T, row.names = 1)Helper function
Helper function for data wrangling. Combines PBMC cell types, convert correlations to 1−PCC, and build three long tables (library size, peak means, sparsity).
Show code
# ---- Helpers ---------------------------------------------------------------
prep_metrics <- function(dfs, invert_cols = 10:12, exclude_sim = "simATAC", order = c('simCAS','simPIC(w)','simPIC(g)'), round_peakmean = TRUE, log_libsize = FALSE) {
# bind & invert correlations (or whichever metrics live in invert_cols)
df <- do.call(rbind, dfs)
df[, invert_cols] <- 1 - df[, invert_cols]
# Common factor order
df$simulator <- factor(df$simulator, levels = order)
# Build long-format frames for each metric group
build_long <- function(cols, var_levels, round_first4 = FALSE, maybe_log_first3 = FALSE) {
sub <- df[, c(cols, which(colnames(df) == "simulator"))]
if (maybe_log_first3) sub[, 1:3] <- if (isTRUE(log_libsize)) log(sub[, 1:3]) else sub[, 1:3]
if (round_first4) sub[, 1:4] <- round(sub[, 1:4], 2)
long <- reshape2::melt(sub)
long <- long[long$simulator != exclude_sim, ]
long$variable <- factor(sub("_.*", "", long$variable), levels = var_levels)
long$simulator <- factor(long$simulator, levels = order)
long
}
var_levels <- c("MAD","MAE","RMSE","Correlation")
list(
lib = build_long(cols = c(1,4,7,10,13), var_levels = var_levels, maybe_log_first3 = TRUE),
mean = build_long(cols = c(2,5,8,11,13), var_levels = var_levels, round_first4 = round_peakmean),
spars = build_long(cols = c(3,6,9,12,13), var_levels = var_levels)
)
}
make_panels <- function(long_df, fill_colors = c("#984EA3","#4DAF4A","#377EB8")) {
lapply(unique(long_df$variable), function(var) {
ggplot(long_df[long_df$variable == var, ],
aes(x = simulator, y = value, fill = simulator, colour = simulator)) +
geom_boxplot(width = 0.8, size = 0.6, alpha = 0.3, position = position_dodge2(0.5)) +
geom_jitter(size = 1, alpha = 0.5) +
ggtitle(var) +
scale_fill_manual(values = fill_colors) +
scale_color_manual(values = fill_colors) +
theme_bw() +
theme(
legend.position = "none",
axis.text = element_text(size = 8, color = "black"),
axis.title = element_blank(),
plot.title = element_text(hjust = 0.5, vjust = 0, size = 6),
panel.grid.minor = element_blank(),
panel.grid.major.x = element_blank()
) +
coord_flip()
})
}
row_lab <- function(txt, size = 11, face = "bold") {
cowplot::ggdraw() +
cowplot::draw_label(
txt, x = 0.98, y = 0.5, hjust = 1, vjust = 0.5,
fontface = face, size = size
) +
ggplot2::theme_void()
}
section_lab <- function(txt, size = 12, rotate = TRUE) {
cowplot::ggdraw() +
cowplot::draw_label(
txt, x = 0.5, y = 0.5, hjust = 0.5, vjust = 0.5,
fontface = "bold", size = size,
angle = if (rotate) 90 else 0
) +
ggplot2::theme_void()
}PBMC data formatting using helper function
Adding a simualtor column
Show code
# ---- PBMC ------------------------------------------------------------------
# Ensure simulator column exists first (you already do this)
pbmc10k_cd4memory$simulator <- rownames(pbmc10k_cd4memory)
pbmc10k_cd4naive$simulator <- rownames(pbmc10k_cd4naive)
pbmc10k_cd8effector$simulator <- rownames(pbmc10k_cd8effector)
pbmc10k_cd14mono$simulator <- rownames(pbmc10k_cd14mono)
pbmc10k_cd8naive$simulator <- rownames(pbmc10k_cd8naive)
pbmc10k_bcell$simulator <- rownames(pbmc10k_bcell)
pbmc10k_nkdim$simulator <- rownames(pbmc10k_nkdim)
pbmc_lists <- prep_metrics(
dfs = list(pbmc10k_cd4memory, pbmc10k_cd4naive, pbmc10k_cd8effector, pbmc10k_cd14mono,
pbmc10k_cd8naive, pbmc10k_bcell, pbmc10k_nkdim),
invert_cols = 10:12,
order = c('simCAS','simPIC(w)','simPIC(g)'),
round_peakmean = TRUE,
log_libsize = FALSE # set TRUE to log-transform MAD/MAE/RMSE panels
)
rename_corr <- function(df) {
df$variable <- as.character(df$variable)
df$variable[df$variable == "Correlation"] <- "1-PCC"
df$variable <- factor(df$variable, levels = c("MAD","MAE","RMSE","1-PCC"))
df
}
pbmc_lists <- lapply(pbmc_lists, rename_corr)
pbmc10k_lib_list <- make_panels(pbmc_lists$lib)
pbmc10k_peakmean_list <- make_panels(pbmc_lists$mean)
pbmc10k_sparsity_list <- make_panels(pbmc_lists$spars)Satpathy data formatting using helper function
Adding a simulator column same as PBMC data
Show code
# ---- Satpathy --------------------------------------------------------------
satpathy_monocytes$simulator <- rownames(satpathy_monocytes)
satpathy_naivecd8tcell$simulator <- rownames(satpathy_naivecd8tcell)
satpathy_regtcell$simulator <- rownames(satpathy_regtcell)
satpathy_cd34$simulator <- rownames(satpathy_cd34)
sat_lists <- prep_metrics(
dfs = list(satpathy_monocytes, satpathy_naivecd8tcell, satpathy_regtcell, satpathy_cd34),
invert_cols = 10:12,
order = c('simCAS','simPIC(w)','simPIC(g)'),
round_peakmean = TRUE,
log_libsize = FALSE
)
sat_lists <- lapply(sat_lists, rename_corr)
satpathy_lib_list <- make_panels(sat_lists$lib)
satpathy_peakmean_list <- make_panels(sat_lists$mean)
satpathy_sparsity_list <- make_panels(sat_lists$spars)Generating Figure 3
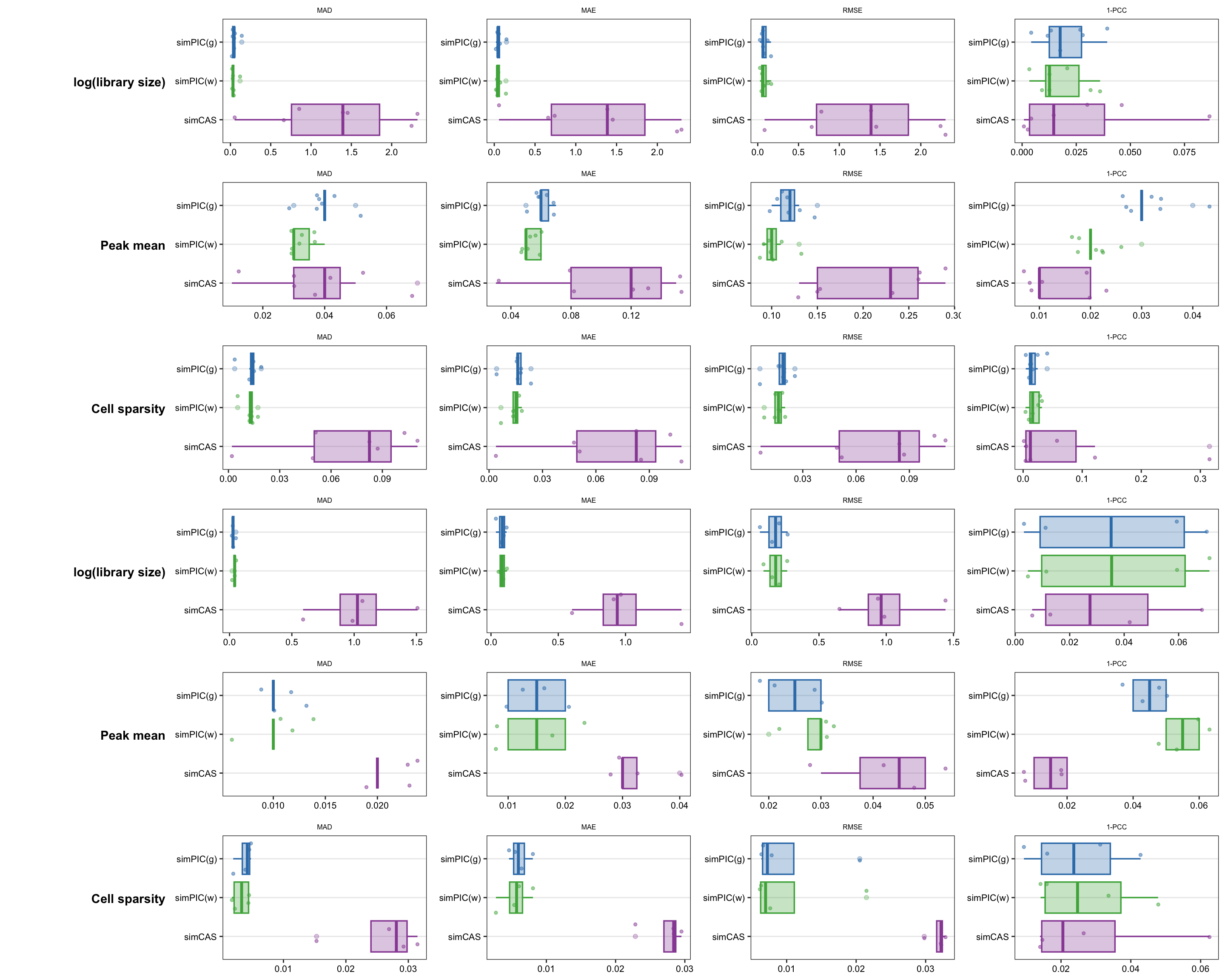
First three rows are for PBMC10k and last three are for Satpathy.
Show code
# ---- Build panels as you already do -----------------------------------------
pbmc10k_lib_list <- make_panels(pbmc_lists$lib)
pbmc10k_peakmean_list <- make_panels(pbmc_lists$mean)
pbmc10k_sparsity_list <- make_panels(pbmc_lists$spars)
satpathy_lib_list <- make_panels(sat_lists$lib)
satpathy_peakmean_list <- make_panels(sat_lists$mean)
satpathy_sparsity_list <- make_panels(sat_lists$spars)
# Main 6 rows × 4 cols grid
main_grid <- cowplot::plot_grid(
plotlist = list(
pbmc10k_lib_list[[1]], pbmc10k_lib_list[[2]], pbmc10k_lib_list[[3]], pbmc10k_lib_list[[4]],
pbmc10k_peakmean_list[[1]], pbmc10k_peakmean_list[[2]], pbmc10k_peakmean_list[[3]], pbmc10k_peakmean_list[[4]],
pbmc10k_sparsity_list[[1]], pbmc10k_sparsity_list[[2]], pbmc10k_sparsity_list[[3]], pbmc10k_sparsity_list[[4]],
satpathy_lib_list[[1]], satpathy_lib_list[[2]], satpathy_lib_list[[3]], satpathy_lib_list[[4]],
satpathy_peakmean_list[[1]],satpathy_peakmean_list[[2]],satpathy_peakmean_list[[3]],satpathy_peakmean_list[[4]],
satpathy_sparsity_list[[1]],satpathy_sparsity_list[[2]],satpathy_sparsity_list[[3]],satpathy_sparsity_list[[4]]
),
ncol = 4, align = "v"
)
# Left-side row labels (PBMC rows 1–3, Satpathy rows 4–6)
row_lab <- function(txt, size = 11) {
cowplot::ggdraw() +
cowplot::draw_label(txt, x = 0.98, y = 0.5, hjust = 1, vjust = 0.5, fontface = "bold", size = size) +
theme_void()
}
left_labels <- cowplot::plot_grid(
row_lab("log(library size)"),
row_lab("Peak mean"),
row_lab("Cell sparsity"),
row_lab("log(library size)"),
row_lab("Peak mean"),
row_lab("Cell sparsity"),
ncol = 1, rel_heights = rep(1, 6)
)
# Combine labels + main grid
combined_with_labels <- cowplot::plot_grid(
left_labels, main_grid,
ncol = 2, rel_widths = c(0.16, 1), align = "h"
)
combined_with_labels